Resources
We release open-source software, models, and datasets associated with our research publications.
Featured

Tools for deploying our spatial aging clock models (trained on mouse brain data) to a single-cell datasets (both spatial or non-spatial).
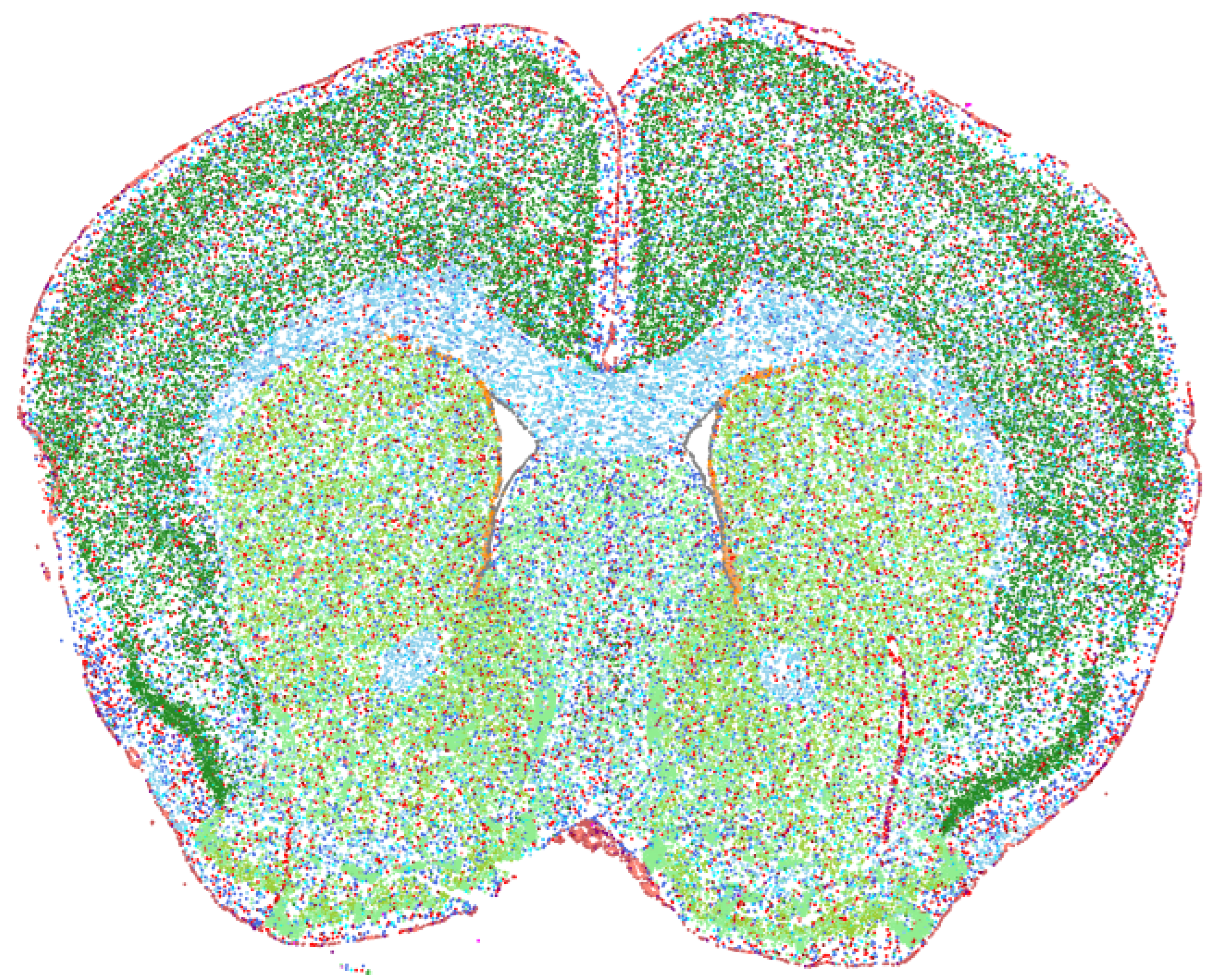
This Zenodo repository contains AnnData objects containing the processed MERFISH spatial transcriptomics datasets for mouse brain aging, experiments with exercise, and experiments with partial reprogramming.
TISSUE (Transcript Imputation with Spatial Single-cell Uncertainty Estimation) provides tools for estimating well-calibrated uncertainty measures for gene expression predictions in single-cell spatial transcriptomics datasets and utilizing them in downstream analyses.
SPRITE (Spatial Propagation and Reinforcement of Imputed Transcript Expression) is a meta-algorithm for post-processing spatial gene expression predictions to improve the quality of predictions and their utility in downstream analysis.
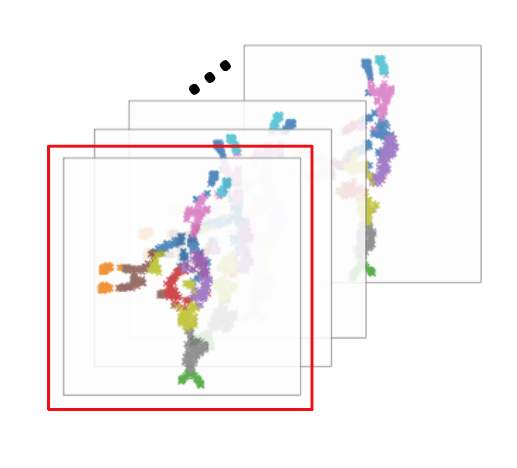
DynamicViz package for generating bootstrap visualizations of high-dimensional data. Great for assessing stability of visualizations and increasing robustness of interpretations.
